Computational Analysis of the Dynamics of Fluorescent RNA Binding Proteins
This project involved analyzing the dynamics of spliceosome nuclear condensates in live fluorescence microscopy videos obtained from the salivary gland nuclei of third-instar Drosophila larvae. My goal was to investigate the effect of the prion-like domain of the transcription factor CLAMP on the behavior and formation of RNA-binding protein condensates involved in alternative splicing across both male and female experimental backgrounds. This research was conducted during my time as an undergraduate in the Larschan Lab at Brown University.
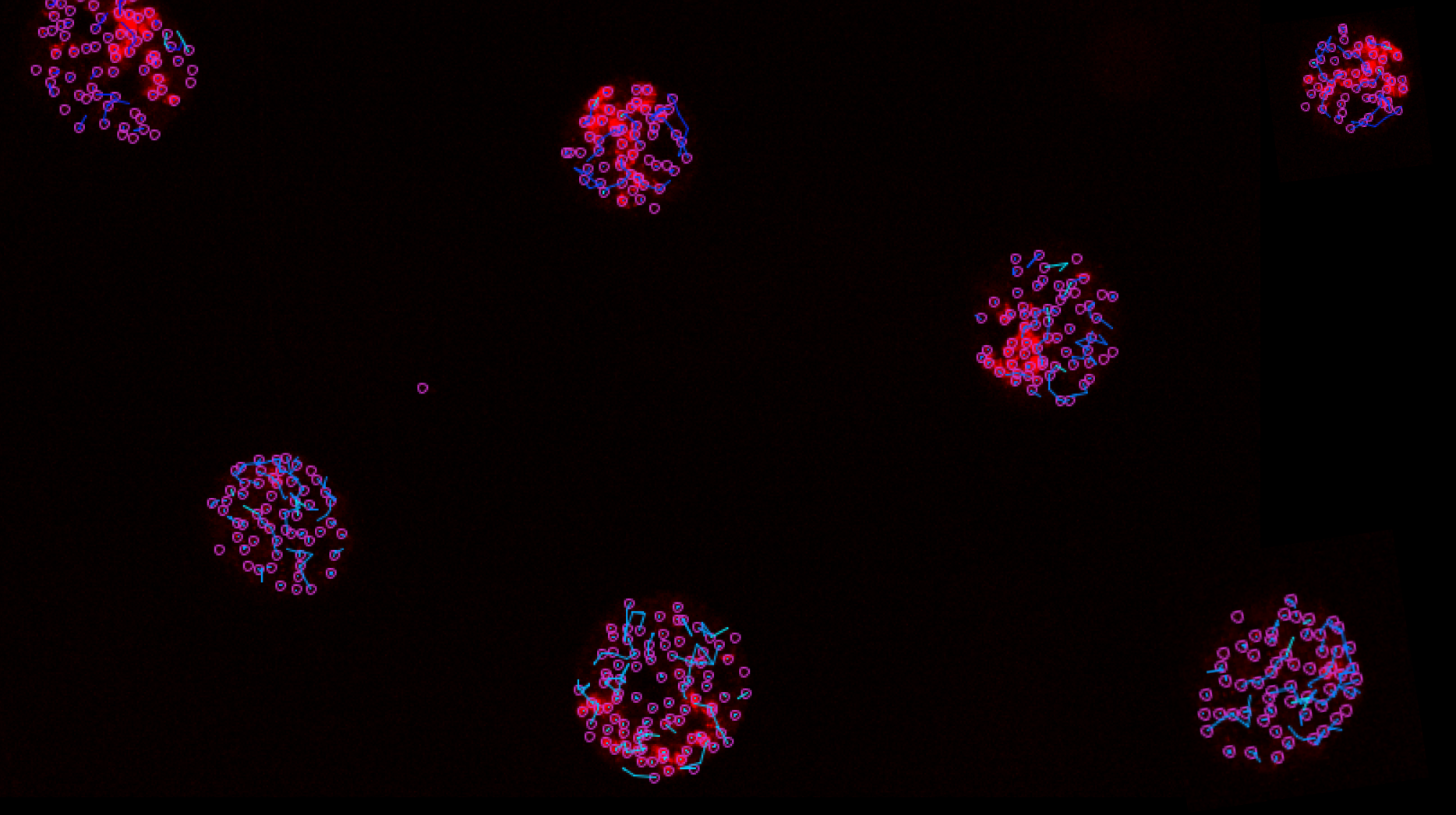
To achieve this, I developed an image processing, segmentation, tracking, and data analysis pipeline using the open-source package TrackIt, which quantified the behavior of these condensates under varying experimental conditions. The image processing script was designed to automate the Fiji image processing tasks and was written as an ImageJ macro (Jython).
More details about this project can be found in my undergraduate honors thesis, linked here: Honors Thesis. Additionally, my work on this project is part of a paper currently available as a preprint: PubMed.
An example track produced by TrackIt. Here we can see a condensate (pink track) move, stop, start moving again, and then possibly merge with another condensate.
Here is an example of results produced from this pipeline: this is a measure of the pooled bound fraction from each experimental background. We can see that in our mutant background, the condensates appear to spend more time “bound” or in the non-mobile phase than in the control backgrounds.

